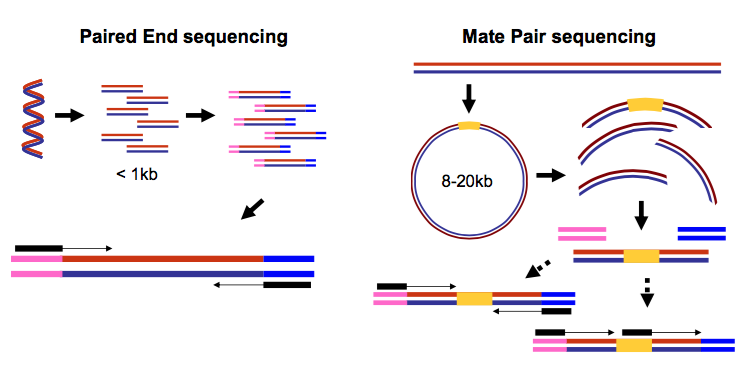
paired end sequencing wikipedia
There is a unique adapter sequence on both ends of the paired-end read labeled Read 1 Adapter and Read 2 Adapter. The inner mate distance between the two reads is.
The latter one is also called mate pair.

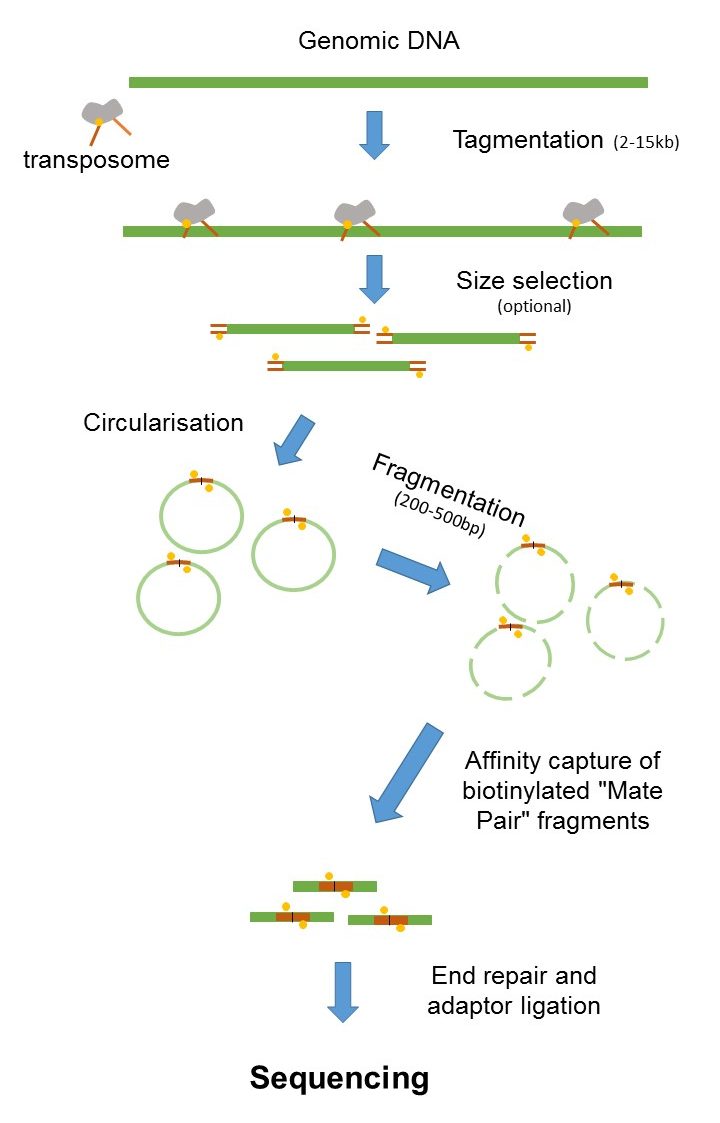
. The figure shows the workflow for mate-pair library preparation for Illumina sequencing. In addition the second part of identifier related to the paired number is always one. The current read length of this technique is 26 bases per amplicon and 13 bases per tag leaving a gap of 45 bases in each tag.
Single-Read Sequencing Technology Paired-end runs sequence both DNA ends for easier analysis of rearrangements novel transcripts and more. For the first test I took some sequence from the human genome hg19 and created two 100 bp reads from this region. It also requires an extremely high sequencing depth of around 5 billion paired-end reads per sample to achieve the resolution of data described by Rao et al.
Die beiden Enden eines Fragments werden auch als mate pairs bezeichnet. An illustrated procedure for. The other problem is with the indexes they are not same some times in an individual file.
To simplify you can differ between two kinds of reads for paired-end sequencing. Paired-end reads are preferable for de novo transcript discovery or isoforms expression analysis as well as to characterise poorly annotated transcriptomes. Diese vier methoden umfassen die endpaar-sequenzierung und deren bioinformatische analyse paired-end mapping die direkte alinierung von dna-sequenzen über die endstellen oder bruchpunkte von cnvs split-read analysis die sequenztiefenanalyse read-depth analysis und die verwendung von bioinformatischen methoden zur.
Now lets get started. This will only go through the first 10000 lines which makes this one-liner much much faster. Below is a list of paired end sequencing words - that is words related to paired end sequencing.
Yes that is my understanding as well. SIPERs are 200800 bp long LIPERs can be longer. Shortinsert pairedend reads SIPERs and long-insert paired-end reads LIPERs.
Described below are two of the most prominent in situ Hi-C based techniques. For those not familiar with paired-end reads check out this post. The words at the top of the list are the ones most associated with paired end sequencing and as you go.
The structure of a paired-end read is described here. Individual reads can be paired together to create paired-end reads which offers some benefits for downstream bioinformatics data analysis algorithms. It has very nice and simple illustrations along with explanations on the terminology used in paired-end sequencing.
The top 4 are. The difference between the two variants is first surprise - the length of the insert. What Are Paired End Reads The Sequencing Center In addition to fragment.
US7601499B2 - Paired end sequencing - Google Patents Paired end sequencing Download PDF Info Publication number US7601499B2. Paired-end tags sequence assembly genetics and sequencingYou can get the definitions of a word in the list below by tapping the question-mark icon next to it. Each sequence is called an end-read or read 1 and read 2 and two reads from the same clone are referred to as mate pairs.
It has been done with illumina paired end sequencing. In short-read sequencing intact genomic DNA is sheared into several million short DNA fragments called reads. But the reads identifiers for the forward and reverse read of one sequence is not match at all.
End-sequence profiling ESP sometimes Paired-end mapping PEM is a method based on sequence-tagged connectors developed to facilitate de novo genome sequencing to identify high-resolution copy number and structural aberrations such as inversions and translocations. Sequencing depth or library size. This is particularly useful if you have paired-ends in separate files as you will get all ones form one and all twos from the other.
Since the beginning of 2013 this preparation has been based on Nextera technology. It has very nice and simple illustrations along with explanations on the terminology used in paired-end. Paired-end sequencing means sequencing both ends of the cDNA fragments and aligning the forward and reverse reads as read pairs Figure 8.
Since the chain termination method usually can only produce reads between 500 and 1000 bases long in all but the smallest clones mate pairs. Die Länge und die beiden Endsequenzen jedes Fragmentes werden in der späteren Assemblierungsphase der Fragmente verwendet. Polony sequencing is generally performed on paired-end tags library that each molecule of DNA template is of 135 bp in length with two 1718 bp paired genomic tags separated and flanked by common sequences.
Single-end runs offer an economical alternative. Single read vs paired end sequencing in RNAseq. The preparation of mate pair libraries is designed to allow classical paired-end sequencing of both ends of a fragment with an original size of several kilobases.
Paired-end sequencing involves sequencing both ends of a fragment and facilitates detection of genomic rearrangements as well as gene fusions and novel transcripts. Paired-end reads are innie and mate pairs are outie Sanger paired ends are generated from a completely different process sequencing the ends of BAC clones and the result is that those paired ends are outie This leads to a lot of confusion when using a mix of technologies or using software that. Several techniques that have adapted the concept of in situ Hi-C exist including Sis Hi-C OCEAN-C and in situ capture Hi-C.
Paired end sequencing wikipedia Monday April 4 2022 Edit. Skip to content ProductsLearnCompanySupportRecommended Links Products Instruments Kits Reagents Selection Tools Software Analysis Services. Specifically the invention provides cloning and DNA manipulation strategies to isolate the two ends of a large target nucleic acid into a single small DNA construct for rapid cloning sequencing or amplification.
Difference Between Whole Genome Sequencing And Next Generation Sequencing Difference Between

What Are Paired End Reads The Sequencing Center

Sequencing Introduction To Sequencing By Synthesis Sbs Youtube
Manipulating Ngs Data With Galaxy Galaxy Community Hub
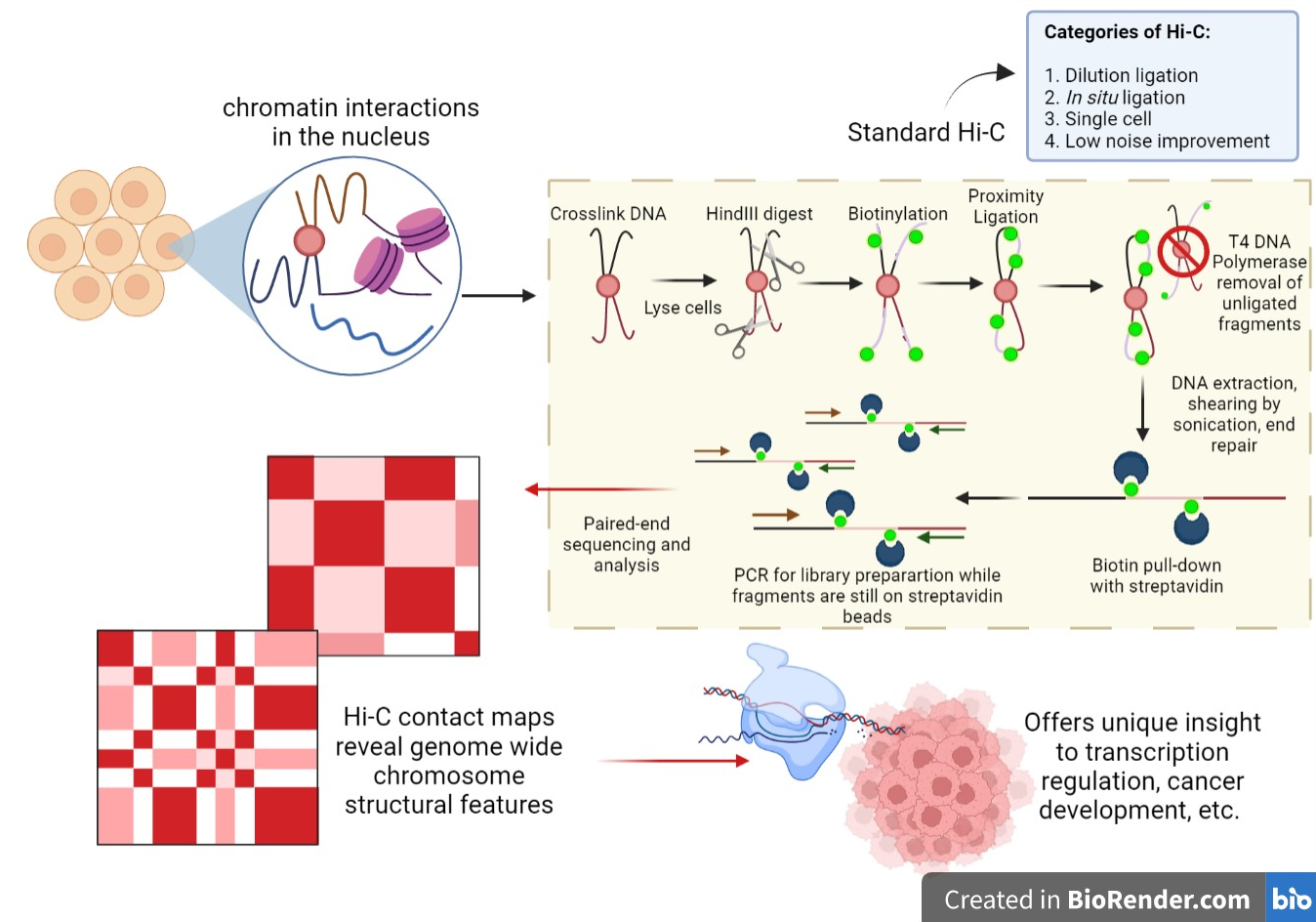
Hi C Genomic Analysis Technique Wikipedia
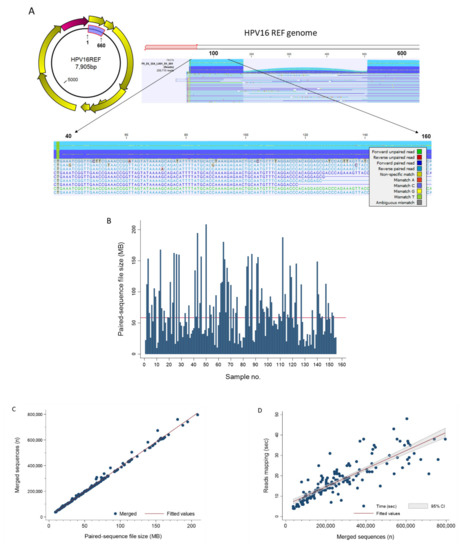
Pathogens Free Full Text Hpv Deepseq An Ultra Fast Method Of Ngs Data Analysis And Visualization Using Automated Workflows And A Customized Papillomavirus Database In Clc Genomics Workbench Html

Rahul Satija Satijalab Twitter

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

Fundamental Principles In Cardiovascular Genetics Sciencedirect

Bacterial Chromosome Replication Biology Lessons Chromosome Biology

What Is Mate Pair Sequencing For

What Are Paired End Reads The Sequencing Center